
, . - decrease / increase font size
DEL - remove font from current set Home - restore font set to initial
Case Studies
Protein Structure Prediction
The laboratories of Karl Freed and Tobin Sosnick use Swift to develop and validate methods to predict protein structure using homology-free approaches.
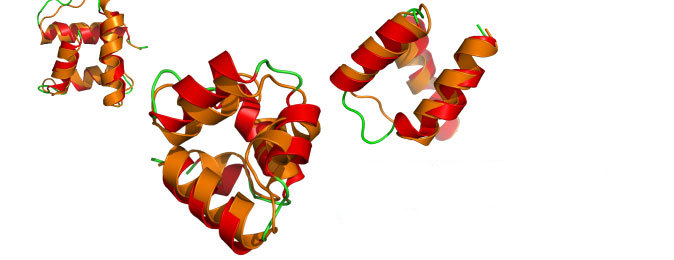
Approach. A. Adhikari (under K. Freed and T. Sosnick) has developed new structure prediction techniques based on Monte Carlo simulated annealing which employ novel, compact molecular representations and innovative “moves” of the protein backbone to achieve accurate prediction with far less computation then previous methods. One of the applications of the method involves rebuilding local regions in protein structures, called “loop modeling”, a problem which the group tackled with considerable success in the CASP protein folding tournament(shown in right). They are now testing further algorithmic innovations using the computational power of Beagle.
Results. The group is now developing a new iterative algorithm for predicting protein structure and folding pathway starting only from the amino acid sequence. In progress, no publications yet from Beagle studies.